|
Hi there! I'm glad you made it to my digital portfolio :) I am a 2nd-year Bioengineering PhD student at University of Pennsylvania, advised by Derek Oldridge. I am interested in developing statistical and AI-driven analytical tools that integrate spatial-omic technologies with conventional pathology imaging to study spatial biology. My goal is to enhance our understanding of the molecular, cellular, and architectural foundations of human health and disease in immunological context . Email / CV / LinkedIn / GitHub / Google Scholar |

|
|
|

|
School of Engineering and Applied Science, University of Pennsylvania Children's Hospital of Philadelphia Sep 2024 - Present | Philadelphia, PA, USA Advisor: Derek Oldridge |

|
Whiting School of Engineering, Johns Hopkins University Sep 2022 - Apr 2024 | Baltimore, MD, USA GPA: 4.0/4.0 Advisor: Denis Wirtz & Ashley Kiemen |

|
Faculty of Applied Science and Engineering, University of Toronto Sep 2017 - Apr 2022 | Toronto, ON, Canada Dean’s Honors List, 2020-2022 U of T Entrance Scholarship, 2017 Thesis Advisor: Eli Sone (2021-2022) Research Advisor: Michael Garton (2021) Research Advisor: Alison McGuigan (2019) |
|
|
|
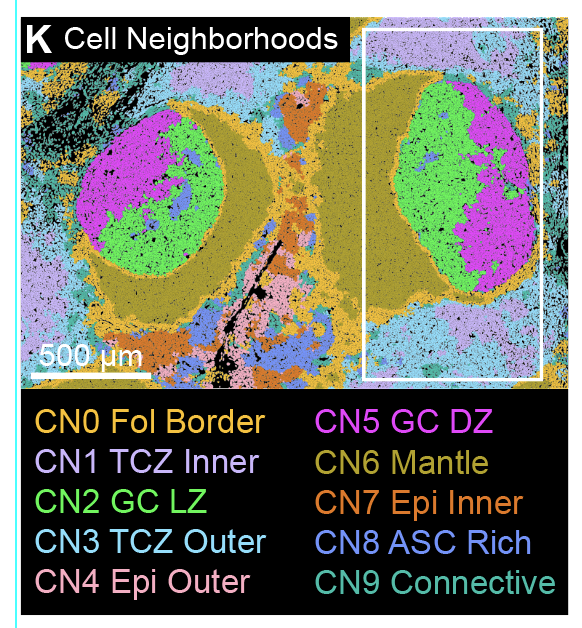
Sam Barnett Dubensky, Yutong Zhu, Molly Gallagher, Kingsley Gideon Kumashie, Tianyu Lu, Jonathan Tedesco, Nina De Luna, Katherine Premo, Yi Qi, Suzanna Rachimi, Emylette Cruz Cabrera, Bria Fulmer, Ijeoma C. Meremikwu, Ashley Carter, Sarah E. Henrickson, Neil Romberg, Amy E. Baxter, Derek A. Oldridge, Laura A. Vella Submitted, 2025 bioRxiv preprint Using multimodal single-cell and spatial transcriptomic profiling of human tonsils and blood, we define distinct CD4 T follicular helper (Tfh) cell states and identify GNG4 as a hallmark of activated, germinal center–associated Tfh cells. GNG4 expression more specifically marks GC-localized Tfh than canonical markers, distinguishing activated GC Tfh from resting, non-GC, Th17-polarized Tfh states. |

|
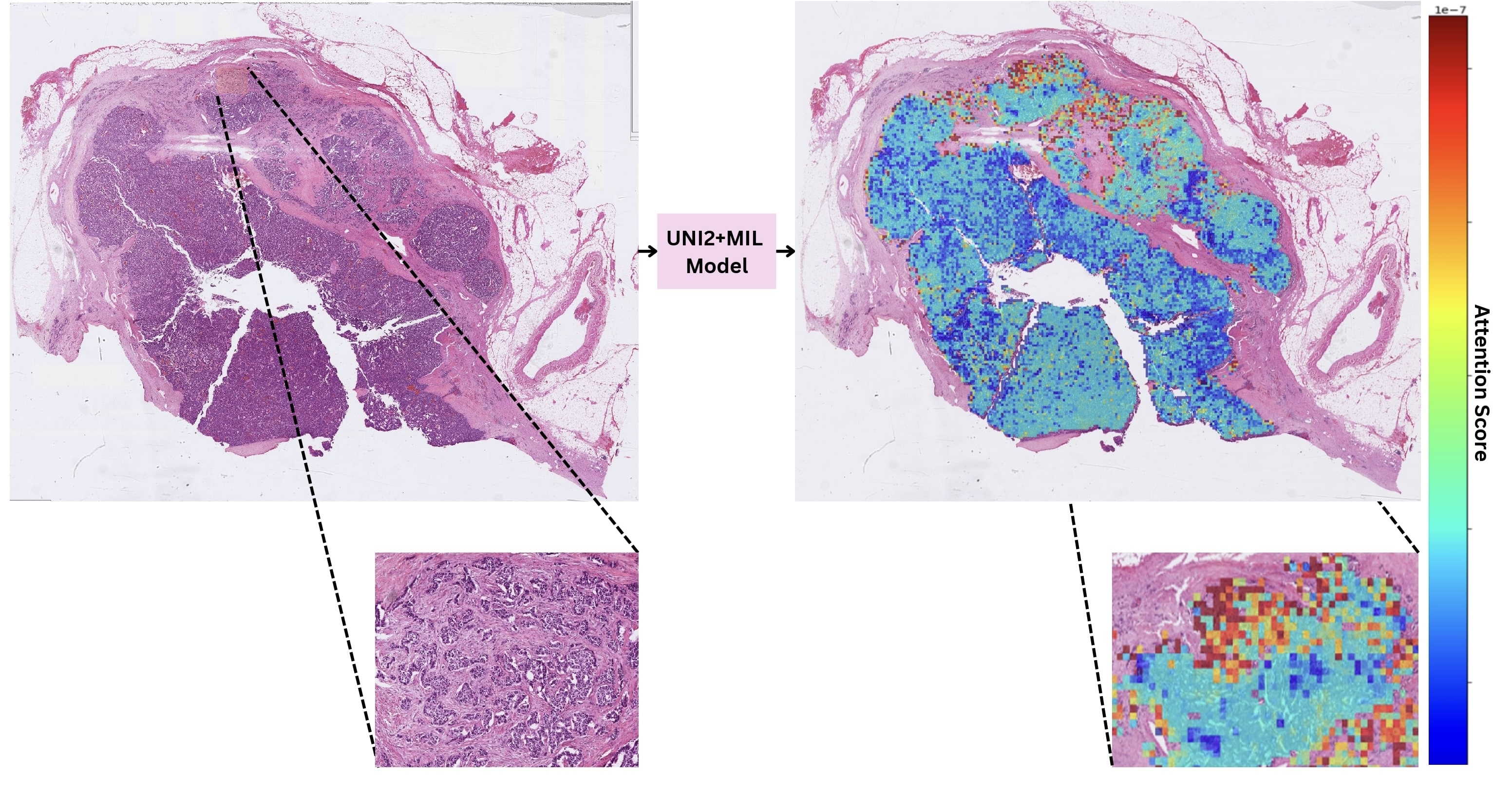
Arrun Sivasubramanian, Lucie S R Dequiedt, Yu Shen, Valentina Matos-Romero, Yutong Zhu, Uwakmfon-Abasi Imoh Ebong, Zoe Gaillard, Ian Reucroft, Chen Mayer, Hyun-Nam Yun, Seung-Mo Hong, Jinho Shin, Atsuko Kasajima, Günter Klöppel, Jin He, Amanda L Blackford, Ralph H. Hruban, Ashley L Kiemen USCAP Annual Meeting (Accepted), 2025 Website We developed an AI-based histopathology model to distinguish long-term disease-free PanNET patients from those who later recurred, using 3,888 H&E images from 926 resections. The pipeline achieved high accuracy (87–89%) across recurrence thresholds and attention maps revealed that small regions, often at invasive edges or vascular invasion—drove prediction. These findings highlight AI as a promising tool for postoperative risk stratification in PanNETs. |
|
Alicia M. Braxton, Ashley L. Kiemen*, Mia P. Grahn, André Forjaz, Jeeun Parksong, Jaanvi Mahesh Babu, Jiaying Lai, lily zheng, Noushin Niknafs, Liping Jiang, Haixia Cheng, Qianqian Song, Rebecca Reichel, Sarah Graham, Alexander I. Damanakis, Catherine G Fischer, Stephanie Mou, Cameron Metz, Julie Granger, Xiao-Ding Liu, Niklas Bachmann, Yutong Zhu, YunZhou Liu, Cristina Almagro-Perez, Ann Chenyu Jiang, Jeonghyun Yoo, Bridgette Kim, Scott Du, Eli Foster, Jocelyn Y. Hsu, Paula Andreu Rivera, Linda C. Chu, Fengze Liu, Elliot K. Fishman, Alan Yuille, Nicholas J. Roberts, Elizabeth D. Thompson, Robert B. Scharpf, Toby C. Cornish, Yuchen Jiao, Rachel Karchin, Ralph H. Hruban, Pei-Hsun Wu, Denis Wirtz*, Laura D. Wood* Nature, 2024 Article The study involves quantitative assessment of human pancreatic intraepithelial neoplasia (PanIN) using CODA, a novel machine-learning pipeline for 3D image analysis that generates quantifiable models of large pieces of human pancreas with single-cell resolution. By leveraging CODA, for the first time, we are able to describe the spatial and genetic multifocality of human PanINs, providing important insights into the initiation and progression of pancreatic neoplasia. |


|
Ashley L. Kiemen, Lucie Dequiedt, Yu Shen, Yutong Zhu, Valentina Matos-Romero, André Forjaz, Kurtis Campbell, Will Dhana, Toby C. Cornish, Alicia M. Braxton, Pei-Hsun Wu, Elliot K. Fishman, Laura D. Wood Denis Wirtz, Ralph H. Hruban American Journal of Surgical Pathology, 2024 Article This study reevaluates the conventional clinical classification of precursor lesions in Pancreatic Ductal Adenocarcinoma (PDAC), namely PanIN (Pancreatic Intraepithelial Neoplasia) and IPMN (Intraductal Papillary Mucinous Neoplasms). It reveals the inadequacy of the traditional classification when assessing high-resolution 3D images. By constructing cellular-resolution 3D maps, the research uncovers instances where some PanINs grow to macroscopic sizes in narrow ducts, meeting the criteria for IPMNs. |
|
Nancy T. Li, Nila C. Wu, Ruonan Cao, Jose L. Cadavid, Simon Latour, Yutong Zhu, Mirjana Mijalkovic, Reza Roozitalab, Natalie Landon-Brace, Faiyaz Notta, Alison P. McGuigan Biomaterials, 2022 Article The study focuses on developing a Scaffold-supported Platform for Organoid-based Tissues (SPOT) platform compatible with generic 96/384 platforms, using 3D design and bioprinting techniques. SPOT enables a 3D heterogeneous environment that mimics the tumor microenvironment, serving as a useful tool to assess the drug sensitivity of patient-derived organoids and can be easily integrated into the drug discovery pipeline. |
|
|
|
Yutong Zhu, Mia P. Grahn, Aanya Kheterpal, Ashleigh Crawford, Denis Wirtz, Ashley L. Kiemen Johns Hopkins Annual ChemBE Research Symposium (Star poster), 2023 Presenter: Yutong Zhu Poster The study involves the construction of 3D, multi-organ volumes of the normal reproductive system of mice at different age groups using CODA, a novel machine-learning pipeline for 3D image analysis, to qualitatively and quantitatively identify major changes to ovary, fallopian, cervix, and vagina morphology and immune infiltration with age. |

|
Yutong Zhu, Angelico Obille, Eli Sone Engineering Science Undergraduate Thesis Presentation, 2022 Presenter: Yutong Zhu Thesis Defense / Thesis Report The study focuses on the distinct adhesion mechanism employed by freshwater mussels. By quantitatively analyzing localization profiles and post-translational modifications of the adhesive proteins using a staining-based approach, this work provides a well-established role model for developing novel bioadhesives such as self-healing materials and advanced coatings in aqueous environments. |

|
Professional Co-op Experience Year, Garton Lab, University of Toronto Yutong Zhu, Aaron Rosenstein, Michael Garton Final Presentation Slides Designed and developed a library of landing pad cell lines targeting pSH231 and AAVS1 safe harbor sites. The library provides an efficient approach for recombinase-mediated cassette exchange with a specific promoter and gene of interest. *Any future work involving the utilization of the landing pad cell lines will credit this work done by Yutong Zhu, Aaron Rosenstein and Michael Garton |
|
|

|
BME489 Biomedical Systems Engineering Design, University of Toronto Final Report / Final Presentation Slides Designed and simulated a device that prepares the subtalar joint for fusion during tibiotalocaneal arthrodesis, which consists of 2 modular components: a cutting tool and a guide tube. |
|
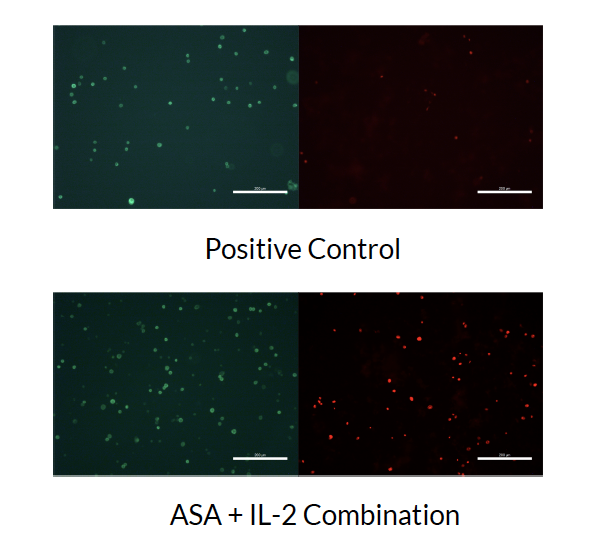
BME346 Biomedical Engineering & Omics Technologies, University of Toronto Final Presentation / Project Proposal Discovered great potential in the combination of platelet inhibition and natural killer (NK) cell enhancement, where platelet inhibition sensitizes tumor cells to NK cell activity and NK enhancement increases cytotoxic effects. |

|
University of Toronto Aerospace Team Demo Engineered C. albicans to express GFP with specific genes and developed a statistical method for quantifying this gene expression following long-time space mission. |
|
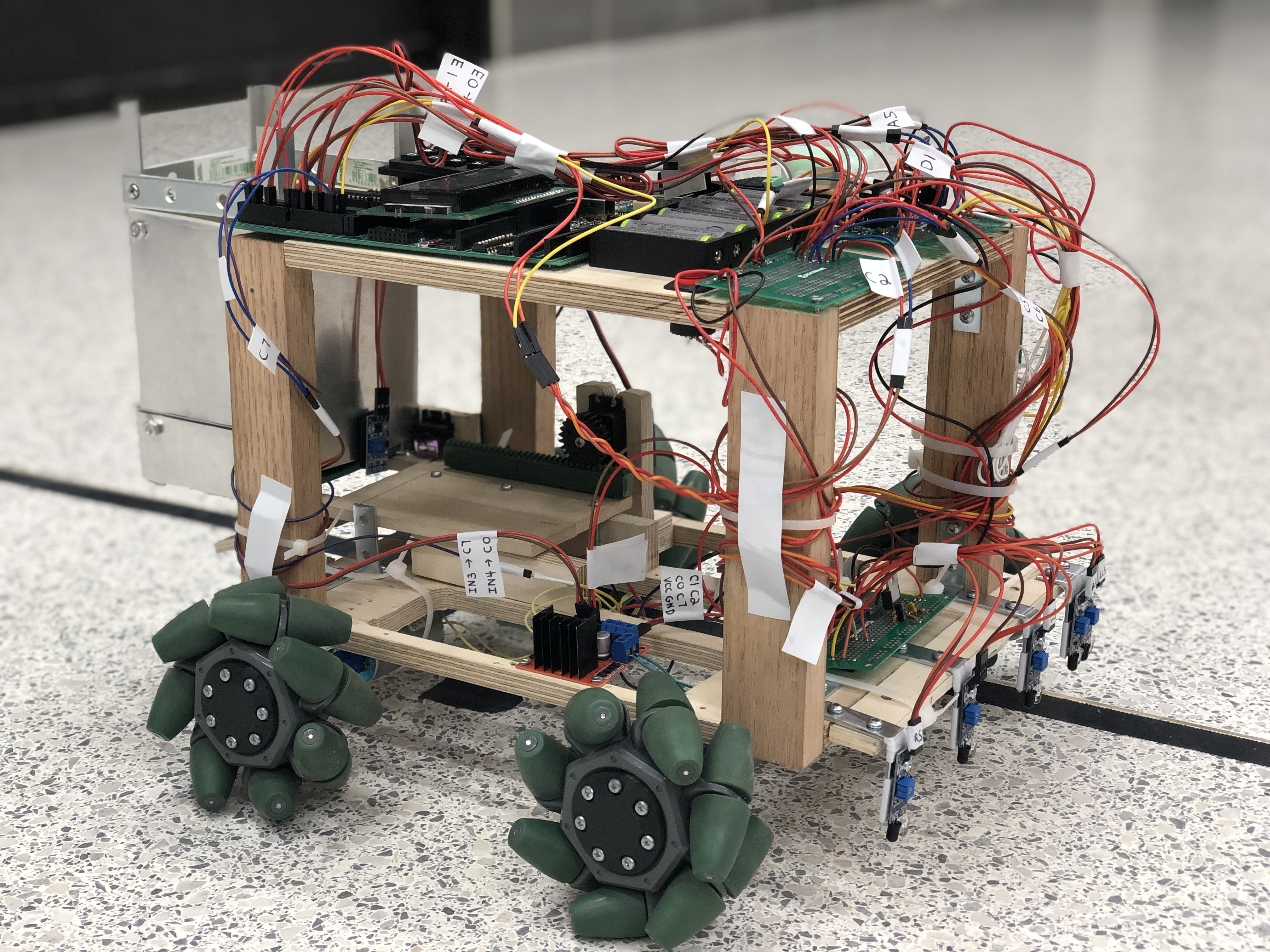
AER201 Engineering Design, University of Toronto Final Report / Demo Designed and fabricated of a fully autonomous mobile robot that can deploy miniature traffic cones based on the detection of "cracks" and "holes". The robot was constructed with omnidirectional mecanum wheels, infrared sensors, as well as an interactive LCD and keyboard user interface. |